The model was based on the rate equations derived from the kinetic model in Figure . All the reactions and parameters used in this computer model were either based on published literatures or measured in M.P.M. lab. For each substrate protein passing through a cycle of refolding process, a probability is assigned for it to be fully refolded. Protein aggregation is not included in our model. This is because of: 1. Lack of quantified data on aggregation; 2. The model is developed to test the property of the DnaK chaperone system at physiologically optimal growing temperatures, where the probability of protein aggregation is only small.
The simulation was based on an improved version of Gillespie's exact stochastic simulation algorithm by Gibson and Bruck, as implemented in E-CELL v3, an open source computer software package for large scale cellular events simulation developed at Keio university (www.e-cell.org). The simulation results were plotted by using GnuPlot (www.gnuplot.info) without any modification. The power fitting was done by Microsoft Excel program.
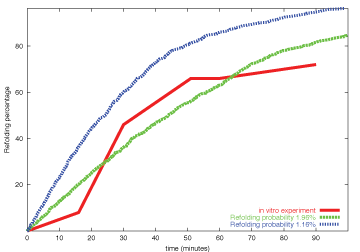
To validate our model, we compared our model results with those published by Schroder. et al. Except this probability value, all other parameters have been reported in literature. As shown in Figure 2, with a refolding probability of 1.16% in a single cycle, results from our model (green) are in consistent with laboratory results (red). A probability value of 1.96% could bring a result with similar dynamics with faster refolding rate (blue).

Figure 2: Model validation