2005öNōxü@ÉXŖŅŗÓɼē╩Ģ±ŹÉÅæ
īżŗåē█æĶ¢╝
üuGenetic Interaction āfü[ā^éŚpéóéĮÉČæ╠ōÓālābāgāÅü[āNŚ\æ¬üv
ōĪ¢{ü@ī░Äm
īcē×ŗ`ÅmæÕŖwü@ÉŁŹ¶üEāüāfāBāAīżŗåē╚ü@ÅCÄméPöNü@
īc£õŗ`ÅmæÕŖwɵÆ[ÉČ¢Įē╚ŖwīżŗåÅŖü@ü@ü@ü@ü@ü@ü@ü@ü@ü@ü@
Abstract
Large-scale
interpretation of genetic interaction networks has begun to reveal the global
organization of Saccharomyces cerevisiae. Recently developed
biological experiment methods bring us to amount of interaction data, but
biological process networks remains obscure. In this research, I detect two
types of cis-network motifs in Saccharomyces cerevisiae gene
networks from genetic interaction data and functional-linkages obtained from
phylogenetic profiling to focus attention on essential gene and showed two
different reason of why essential genes are essential.
Keyword:
phylogenetic profiling, network, genetic interaction, Saccharomyces
cerevisiae
Introduction
One
of the most valuable biological challenges is to interpret post-genomic
networks through recent high-throughput approaches and Saccharomyces
cerevisiae
is appropriate model organism to observe post-genomic networks. The observation
that only ~18% of the genes in Saccharomyces cerevisiae are
essential for viability especially illustrates the capacity of genetic networks
to buffer against genetic perturbation (Giaever, 2002). Direct physical
interactions among yeast proteins are being mapped by systematic two-hybrid and
mass spectrometric characterization of protein complexes (Ito et al., 2001; Uetz
et al.,
2000; Gavin et al., 2002;Ho et al., 2002). Recently
developed methods for the comprehensive identification of synthetic lethal
interactions in Saccharomyces cerevisiae, such as synthetic
genetic arrays (SGA) and synthetic lethal analysis by microarrays (SLAM)
enables large-scale mapping of genetic interactions (Tong et al., 2001; Ooi et
al.,
2003; Tong et al., 2004). Synthetic lethality is an extreme case
in which two single mutations that cause no evident phenotype individually are
lethal in combination. Nature of genetic interaction, it can be a powerful and
widespread tool for establishing functional linkages between genes (Zhang et
al.,
2005; Kelly and Ideker, 2005). And the phylogenetic profiling of protein in
multiple genomes is another high-throughput method for establishing
functional-linkages (Pellegrini et al., 1999).
In
this work, I describe biological process networks discovered from a Saccharomyces
cerevisiae
network by using functional-linkages between genes predicted from the
phylogenetic profiling of protein in multiple genomes and tried to detect two
types of root cis-network motif in recognizing the essential genes.
Materials
and Methods
Data
sources
9226
known physical interaction data and 6389 genetic interaction data were obtained
from Munich Information Center for Protein Sequences (MIPS) (Mewes et al., 2002;
Mewes et al., 2004).
All
of gene information about functions and viability were obtained from the Yeast
Protein Database (YPD)(Hodges et al., 1998).
The
protein-coding sequences of complete genomes were obtained from the NCBI FTP
server.
Phylogenetic
profiling
Phylogenetic
profiles for the 6326 proteins encoded by the genome of Saccharomyces
cerevisiae
were computed by aligning each protein sequence using BLAST search with the
proteins from 99 other fully sequenced genomes. Phylogenetic profiles were
evaluated as if each pair-wised genes coefficient of profiles correlation shows
over 0.7, paired genes were considered as it had functional linkage.
Cis-network
motif
Cis-network
motifs were divided by meaning of role of essential gene into two types. Type I
motif had at least two essential gene and two nonessential genes. Nonessential
genes had synthetic lethality and had functional linkages between one side of
essential genes. Essential genes were also had functional linkages (Figure 1A).
Type
II motif had at least one essential gene and four nonessential genes. Essential
gene had an even number of functional linkages to nonessential genes and pair
of nonessential genes which had functional linkages to essential gene also had
synthetic lethality (Figure 1B).
A B
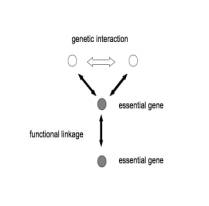
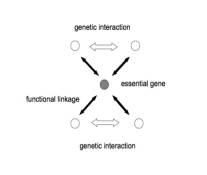
Figure 1, root motif of cis-network
Results
Functional-linkage
networks were predicted including 13998 functional linkages as edges and 1532
genes in Saccharomyces cerevisiae as nodes. This
functional linkage networks resembles the nonessential genetic networks in that
it has a scale-free topology and most of the interactions were nonoverlapping
with protein-protein interaction (Figure 2).
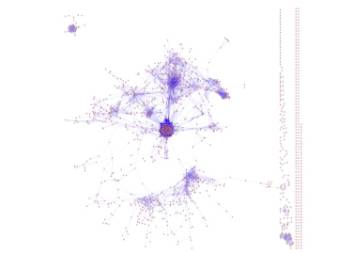
Figure 2, Predicted
functional linkage networks
4360
motifs of type I network (eg. MYO2-TPM1, MYO2-NUM1, MYO2-USO1 as functional
linkage ; TP1-NUM1as genetic interaction; Figure 3A) and 3 motifs of type II
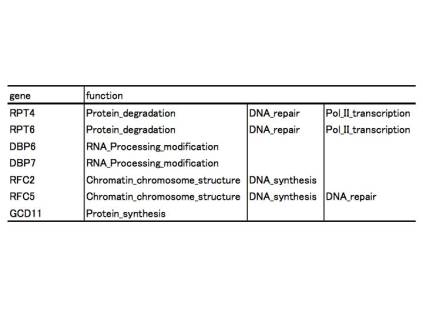
network (eg. HSP60-CCT2, HSP60-CCT4, HSP60-EFT1, HSP60-EFT2 as functional
linkage; CCT2-CCT4, EFT1-EFT2 as genetic interaction, Figure 3B, GCD11-RPT4,
GCD11-RPT6, GCD11-DBP7, GCD11-DBP6, GCD11-RFC2, GCD11-RFC5 as functional
linkage; RPT4-RPT6, DBP6-DBP7, RFC2-RFC5 as genetic interaction, Figure 4) were
detected. Each network node was annotated by use of YPD data (Table 1A, Table
1B, Table 2).
A B
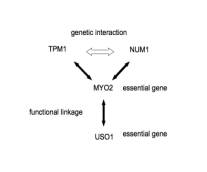
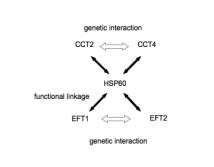
Figure 3, network motif in Saccharomyces
cerevisiae
B A Table 1, gene function
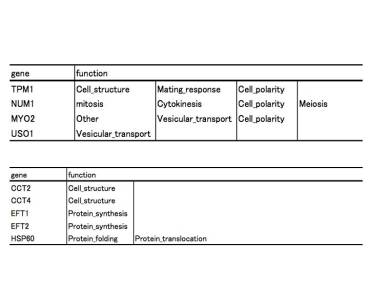
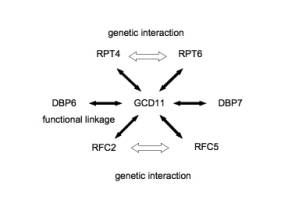
Figure 4, rare network motif in Saccharomyces
cerevisiae
Table 2, gene function for GCD11
network
Discussions
Phylogenetic
profiling and genetic interaction is definitely powerful tool to predict
biological process networks. In the biological process networks predicted by
phylogenetic profiing, essential genes have a important role to maintain
networks. Genetic interaction that it represents synthetic lethality is helpful
to interpret post-genomic networks. In this work, root of cis-network motifs is
detected such as type I and type II motifs. Type I networks like MYO2 network
(Figure 3A) is found in large numbers. The feature of type I network is considered
that essential gene mediates alternative pathway and functional linkage between
essential gene and another is thought to be important in this network motif.
For example, TPM1 and NUM1 has genetic interaction and also has same function
like Cell_polarity (Table 1A), MYO2 which is essential for cell has functional
linkage to TPM1 and MUM1, also has the same function. It shows that MYO2 play a
critical role and edge between MYO2 and USO1 may converge to these pathways.
Central essential gene of type I have hub role in the network, and large number
of existence of type I network in functional linkage networks means that there
is a tendency that essential gene have hub function like protein-protein
interaction networks.
Type
II network like HSP60 networks (Figure 3B) mean bottleneck of biological
process networks. CCT2 to CCT4 thorough HSP60 pathway and EFT1 to EFT2 thorough
HSP60 pathway is different biological process (Table 1B). This types of network
can detect few, this result represent that type II motif is rare because
overconcentration of biological process at one gene is too risky for the cell
to survive. But undue concentration of biological process network is exists
like GCD11 motif (Figure 4). GCD11 networks have several important roles (Table
2), so some sort of biological mechanism for protection of GCD11 may exist.
In
this study, we found cis-network motif in Saccharomyces cerevisiae and interpret
implication of two types of essential genes.
Acknowledgement
I
would like to thank Dr. Rintaro Saito for insightful suggestions and comments.
I also thank Noriyuki Kitagawa for significant discussion. I gratitude for
Prof. Masaru Tomita gives me the opportunity of this research.
References
1. Giaever,G.,
Chu,A.M., Ni,L., Connelly,C., Riles,L., Veronneau,S., Dow,S., Lucau-Danila,A.,
Anderson,K., Andre,B. et al. (2002) Functional profiling of the Saccharomyces
cerevisiae
genome. Nature, 418, 387-391.
2. Ito,T.,
Chiba,T., Ozawa,R., Yoshida,M., Hattori,M. and Sakaki,Y. (2001) A comprehensive
two-hybrid analysis to explore the yeast protein interactome. Proc.Natl
Acad. Sci. USA, 98,4569-4574.
3. Uetz,P.,
Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D.,
Narayan,B., Srinivasan,M., Pochart,P. et al. (2000) Acomprehensive
analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature, 403, 623-627.
4. Gavin,A.C.,
Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M.,
Michon,A.M., Cruciat,C.M. et al. (2002) Functional organization of the
yeast proteome by systematic analysis of protein complexes. Nature, 415, 141-147.
5. Ho,Y.,
Gruhier,A., Heilbut,A., Bader,G.D., Mooore,L., Adams,S.L., Millar,A.,
Taylor,P., Bennett,K., Boutiller,K. et al. (2002) Systematic
identification of protein complexes in Saccharomyces cerevisiae by mass
spectrometry. Nature, 415, 180-183.
6. Tong,A.H.Y.,
Evangelista,M., Parsons,A.B., Xu,H., Bader,G.D., Page,N., Robinson,M.,
Raghibizadeh,S., Hogue,C.W., Busseey,H. et al. (2001) Systematic
genetic analysis with ordered arrays of yeast deletion mutants. Science, 291, 2364-2368.
7. Ooi,S.L.,
Shoemaker,D.D. and Boeke,J.D. (2003) DNA helicase gene interaction network
defined using synthetic lethality analyzed by microarray. Nat Genet, 35, 277-286.
8. Tong,A.H.Y.,
Lesage,G., Bader,G.D., Ding,H., Xu,H., Xin,X., Young,J., Berriz,G.F., Brost,R.,
Chang,M. et al. (2004) Global mapping of the yeast genetic
interaction network. Science, 303, 808-813.
9. Zhang,L.V.,
King,O.D., Wong,S.L., Goldberg,D.S., Tong,A.H., Lesage,G., Andrews,B.,
Bussey,H. and Roth,F.P. (2005) Motifs, themes and thematic maps of an
integrated Saccharomyces cerevisiae interaction network. J
BIol,
4,in
press.
10. Kelley,R. and
Ideker,T. (2005) Systematic interpretation of genetic interactions using
protein networks. Nat Biotechnol, 23,561-566
11. Pellegrini,M.,
Marcotte,E., Thompson,M., Eisenberg,D. and Yeates,T. (1999) Assigning protein
functions by comparative genome analysis: protein phylogenetic profiles. Proc.
Natl Acad. Sci. USA, 96, 4285-4288.
12. Mewes,H.,
Frishman,D., Guldener,U., Mannhaupt,G., Mayer,K., Mokrejs,M., Morgenstern,B.,
Munsterkotter,M., Rudd,S. and Weil,B. (2002) MIPS: a database for genomes and
protein sequences. Nucleic Acids Res., 30, 31-34.
13. Mewes,H.W.,
Amid,C., Arnold,R., Frishman,D., Guldener,U. Mnnhaupt,G., Munsterkotter,M.,
Pagel,P., Strack,N., and Stumpflen,V. et al. (2004) MIPS: analysis
and annotation of proteins from whole genomes. Nucleic Acids Res., 32, 41-44.
14. Hodges,P.E.,
Payne,W.E. and Garrels,J.I. (1998) The Yeast Protein Databbase (YPD): a curated
proteome database for Saccharomyces cerevisiae. Nucleic Acids Res., 26, 68-72.
Recent Presentation
• Annual
Meeting of Molecular Biology Society Japan
ü@December 7-10, 2005.
Fukuoka, Japan
Prediction of biological
process networks in Saccharomyces cerevisiae using phylogenetic profiling and
genetic interaction data
Kenji Fujimoto1,2, Rintaro Saito1, Masaru Tomita1 üi1Inst. Adv. Biosci., Keio
Univ., 2Bioinfo. Prog.
Grad.ü@Sch. Media &
Governance, Keio Univ.üj