Research Achievement Report for Mori Grant
Main research project: GeNESiS: gene network evolution simulation software
Researcher: Anton Kratz (D3)
Affiliation: Keio University, Graduate School of Media and Governance
We developed GeNESIS: gene network evolution simulation software, an interactive application for the simulation of gene regulatory networks (GRNs).
The underlying model for GRNs is based on the central dogma of molecular biology. Genes have one or more sites for the binding of transcription factors and/or other co-factors. RNA Polymerase binds to the transcription factor / cofactor complex and transcribes the gene; the RNA Polymerase and transcription factor complex is then released from the gene (which will be accepting new binding factors and can get transcribed again), and the transcript is translated into a protein. The resulting protein may undergo post-translational modification(s), and may itself be a transcription factor for another gene or even the same gene, within the network.
The evolution of the GRNs is studied by means of a genetic algorithm - it is important to distinguish the "evolution" of the genetic algorithm (which is an optimization technique for the algorithm), and the evolution of the GRNs. The genetic algorithm optimizes the GRNs, but the evolution of the algorithm is of course completely separate to the evolution of the GRNs. Mating occurs at random, and mutations lead to individuals which are unfit and die out, or which optimize a (user-selected) optimization parameter. For example, the user can define fitness as an increase in biomass; in that case, individuals which decrease biomass die out, while individuals which increase the biomass propagate. The underlying GRN will then optimize towards an increase of biomass.
GeNESiS is an interactive, graphical front-end for an implementation of this model that has been developed in [1]. With GeNESiS, the user can select the optimization parameters that define fitness, set the number of genes, protein, inputs (binding sites), set parameters describing mutation rate, etc. The simulation of the GRNs can then be viewed in a movie-like fashion, with one frame showing one generation in a graph representation (Fig. 1). The graph can be visualized in different layouts, and the "movie" showing the evolution process can be saved together with the simulation itself. It is also possible to select individual generations (timepoints) within the movie, and then simulate this GRN (Fig. 2).
We published an article describing this research [2].
The project homepage is at http://genomics.iab.keio.ac.jp/genesis.html
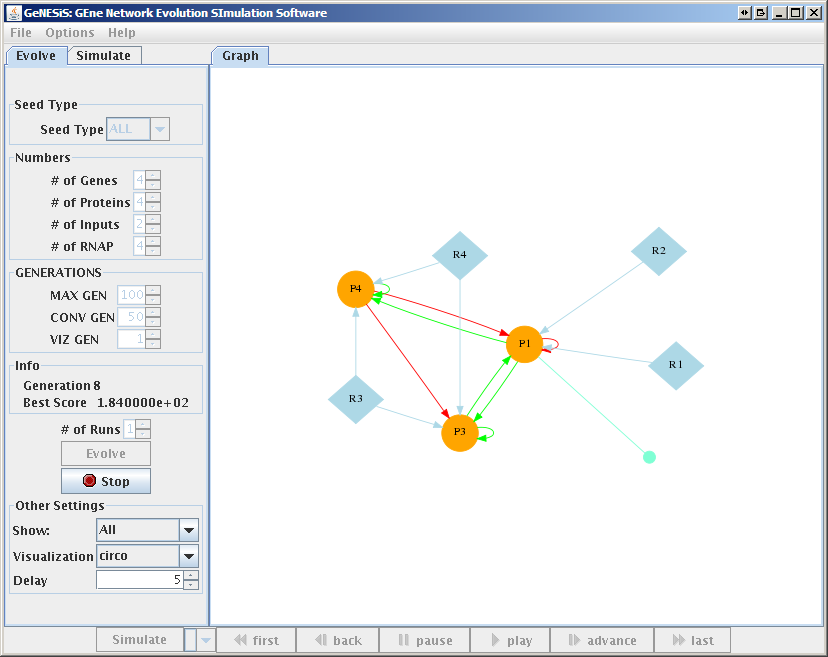
Figure 1: Evolution Window
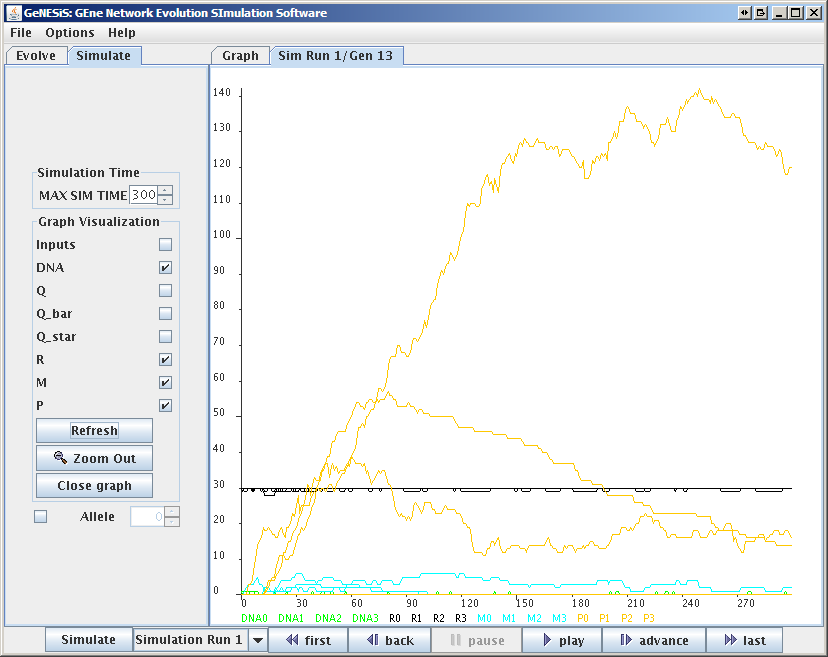
Figure 2: Simulation Window
Literature
[1] Arun Krishnan, Alessandro Giuliani and Masaru Tomita:
"Evolution of Gene Regulatory Networks: Robustness as an emergent property of evolution"
Physica A 387:2170-2186
[2] Anton Kratz, Masaru Tomita, Arun Krishnan:
"GeNESiS: gene network evolution simulation software"
BMC Bioinformatics, Vol. 9, No. 1. (2008)
[Highly accessed, Open Access]