Fig.1: Characterization of five novel sRNAs encoded in the prophages.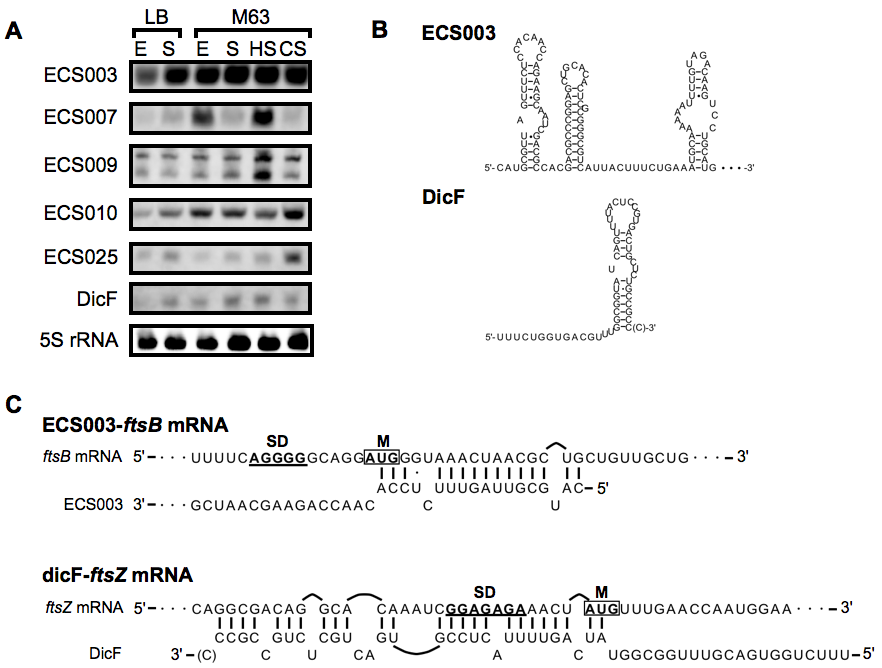
2009年度 森基金 成果報告書
研究課題名: 新規低分子RNAの網羅的予測及び機能解明
新原 温子
政策・メディア研究科 先端生命科学専攻 修士課程1年
Deep sequencing of low molecular weight RNA in Escherichia coli reveals numerous small RNAs transcribed from various region including prophages
Atsuko Shinhara
Systems Biology Program, Graduate School of Media and Governance, Keio University
Research Progress
Backgrounds
Recent studies have suggested that numerous small RNA (sRNA) molecules act as antisense riboregulators in a variety of organisms. In Escherichia coli (E.coli), a large number of sRNAs have been detected using various methods. However, low abundance and/or very small RNAs are difficult to detect by traditional approaches. Consequently, the comprehensive view of the regulation mechanisms through such sRNAs is yet to be clarified.
Results and Discussion
In this study, to focus on these previously unnoticed sRNAs, we utilized Illumina/Solexa deep sequencer for analysis of low molecular weight RNA prepared from E.coli cells grown to late exponential phase. As a result, 6,079 novel transcriptional regions were mapped on the E.coli K12 genome. Then, we selected 160 novel transcriptional regions (≧50 nt) with putative promoters as sRNA candidates; 78 of them were located in intergenic regions and 82 of them were located in cis-antisense strand of the annotated regions. In addition, 10 intergenic sRNAs and four cis-antisense sRNAs were also confirmed by northern blot analysis. Interestingly, transcriptional expressions of four sRNAs in prophage regions were induced by heat-shock or cold-shock stress (Fig.1-A). Furthermore, bioinformatics approach suggested that ECS003 sRNA could regulate ftsB mRNA associated with cell division (Fig.1-B,C).
Fig.1: Characterization of five novel sRNAs encoded in the prophages.
(A) Northern blot analysis of five novel sRNAs encoded in the prophages. Total RNA (20 μg per lane) was isolated from E.coli cells grown to exponential (E) or stationary (S) phase in LB medium or modified M63 minimal medium and from cells subjected to heat shock (HS) or cold shock (CS). Total RNA was also isolated from Δhfq mutant strain grown to exponential phase (Δhfq). 5S rRNA expression was also shown as a loading control. (B) The putative RNA secondary structure of ECS003 sRNA encoded in prophage e14 and the known RNA secondary structure of dicF sRNA encoded in Qin prophage (Faubladier et al., 1990). (C) The base pairing between ECS003 sRNA and ftsB mRNA was predicted using TargetRNA (Tjaden, 2008) with default parameters. The known base pairing between dicF sRNA and ftsZ mRNA (Tetart and Bouche, 1992), were also shown at the bottom. SD: Shine-Dalgarno sequence. M: initiator methionine codon.
Conclusions
In conclusion, we revealed large number of previously unrecognized sRNA transcriptions in E. coli, using deep sequencer. Furthermore, our data will provide a deeper understanding of sRNA regulation in the cell (Fig.2).
Fig.2: Escherichia coli small RNA Browser. Three screenshots were shown as examples of the Escherichia coli small RNA Browser (ECSBrowser). The upper panel showed the overview of the all non-coding RNAs encoded in the E.coli genome; sRNA candidates (red), known sRNAs (pink), tRNAs (green) and rRNAs (yellow). The middle panel magnified the entire prophage e14 regions. The lower panel focused on the ECS003 sRNA. ECS003 sRNA was encoded on the cis-antisense strand of the ymfM RBS. (a) Consensus sequences; promoter (arrow), terminator (vertical line) and RBS (rhombus). (b) Transcriptional regions; sRNA candidates (red), known transcriptional regions (pink) and fragment of known transcripts (black). (c) Known genes; CDS (blue) and pseudogene (white). (d) Deep sequencing data. (e) Genome sequence. Keywords:Deep sequencing; small RNA; cis-antisense sRNA; Prophage; Escherichia coli Congerence Presentation - Talks in accademic conferences -
◉ 第8回 新しいRNA/RNPを見つける会, Hiyoshi, Kanagawa, Japan
- Poster sessions in accademic conferences -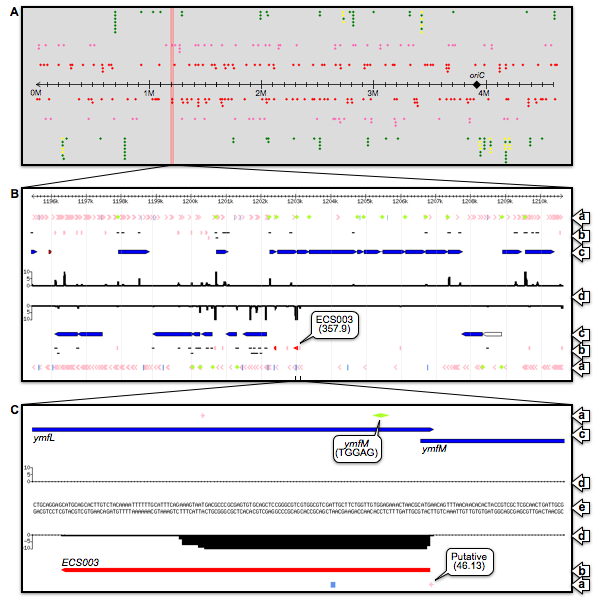
ハイスループットシーケンサを用いた大腸菌におけるSmall RNomics
Deep sequencing analysis suggested novel cis-antisense ncRNAs against ribosomal biding site on mRNAs in Escherichia coli
◉ 日本分子生物学学会32回大会, Yokohama, Kanagawa, Japan
Deep sequencing analysis reveals previously unnoticed small RNAs in Escherichia coli
©Atsuko Shinhara All Rights Reserved Mail:atsuko@sfc.keio.ac.jp